Research
Stochastic modeling of gene expression
Biochemical reactions in living cells are inherently random on the molecular level. But, in spite of that, organisms are able to transform random collisions of molecules driven by thermal fluctuations into higly ordered and complex processes. Mathematical modeling of random processes is a tool which allows us to better understand how disorder is transformed into order inside the living cell.
In [A. Ochab-Marcinek, M. Tabaka, PNAS, 2010] we studied a theoretical model of gene expression in the simplest possible gene regulatory system: a two-step cascade with noncooperative binding of transcription factors. Such a system is deterministically monostable. We have shown that in this system bimodal gene expression is still possible: The reaction of binding of transcription factors to DNA acts as a nonlinear noise filter that transforms the unimodal distribution of transcription factors over the cell population into the bimodal distribution of proteins produced from the regulated gene. We proposed a simple method based on geometric construction that allows one to predict the onset of bimodality.
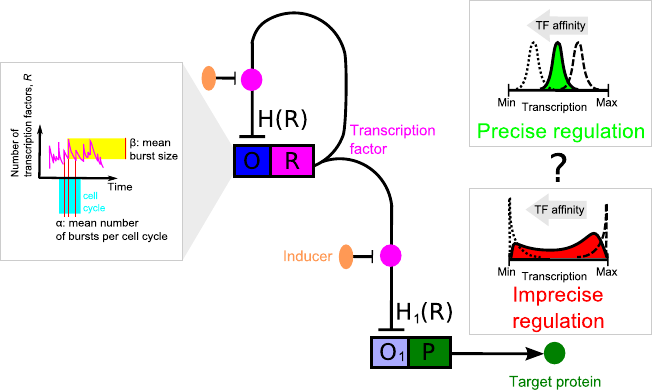
Now, we are extending our study to more complex regulatory motifs to understand what makes their responses to external signals more or less precise with the shapes of protein number distributions ranging from narrow, unimodal, to wide, bimodal. We also take a closer look at the role of bursty protein production and leaky transcription in shaping of the cellular response to signals.
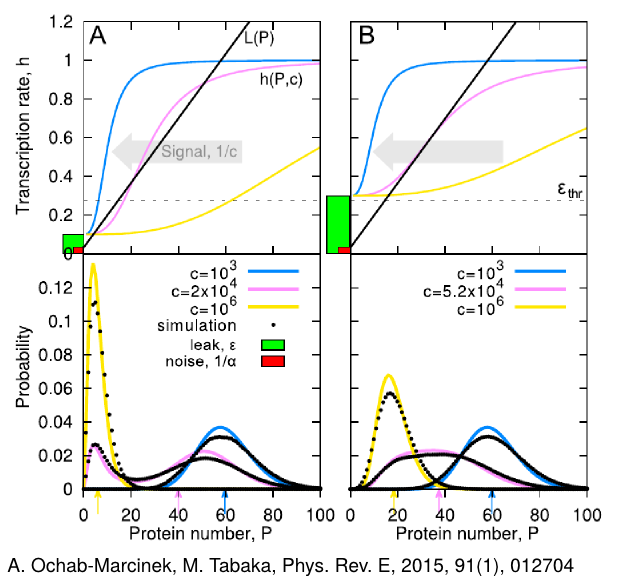
Not all genes are tightly controlled. In nature, many genes show leaky transcription -- some amount of proteins is produced from a given gene even if the gene is in an inactive state. This effect has usually been considered to be unfavourable. In [A. Ochab-Marcinek, M. Tabaka, Phys. Rev. E 2015] we have shown that transcriptional leakage can play an unexpected role in gene expression: A change in its level is relatively easy, as it requires single mutations in gene promoters. It turns out that adjusting the leakage level can change the type of response of a self-regulating gene to external stimulus -- from binary to graded response and vice versa. The same effect obtained by the change of the feedback type (positive/negative) would require many mutations and thus it would be much more difficult to achieve. This suggests that leaky promoters may be so widespread because transcriptional leakage is favored by evolution.
 7.2013-7.2015: Polish Ministry of Science Iuventus Plus grant 0501/IP1/2013/72
7.2013-7.2015: Polish Ministry of Science Iuventus Plus grant 0501/IP1/2013/72
Theoretical study of conditions for precise gene regulation in a 2-gene cascade with autoregulation
Scientific papers published as the result of the project:
- A. Ochab-Marcinek, M. Tabaka, Transcriptional leakage versus noise: A simple mechanism of conversion between binary and graded response in autoregulated genes, Phys. Rev. E, 2015, 91(1), 012704 [arxiv preprint]
- J. Jędrak, A. Ochab-Marcinek, Influence of gene copy number on self-regulated gene expression, J. Theor. Biol., 408 (2016) 222 [arxiv preprint]
- J. Jędrak, A. Ochab-Marcinek, Time-dependent solutions for a stochastic model of gene expression with molecule production in the form of a compound Poisson process, Phys. Rev. E 94 (3) (2016) 032401 [arxiv preprint]
- Hill kinetics as a noise filter: The role of transcription factor autoregulation in gene cascades, Phys. Chem. Chem. Phys., 2017, 19, 22580-22591